
伤口世界
电子邮件地址: 该Email地址已收到反垃圾邮件插件保护。要显示它您需要在浏览器中启用JavaScript。

- 星期四, 04 12月 2025
Circadian Rhythmic Characteristics in Men With Substance Use Disorder Under Treatment. Influence of Age of Onset of Substance Use and Duration of Abstinence




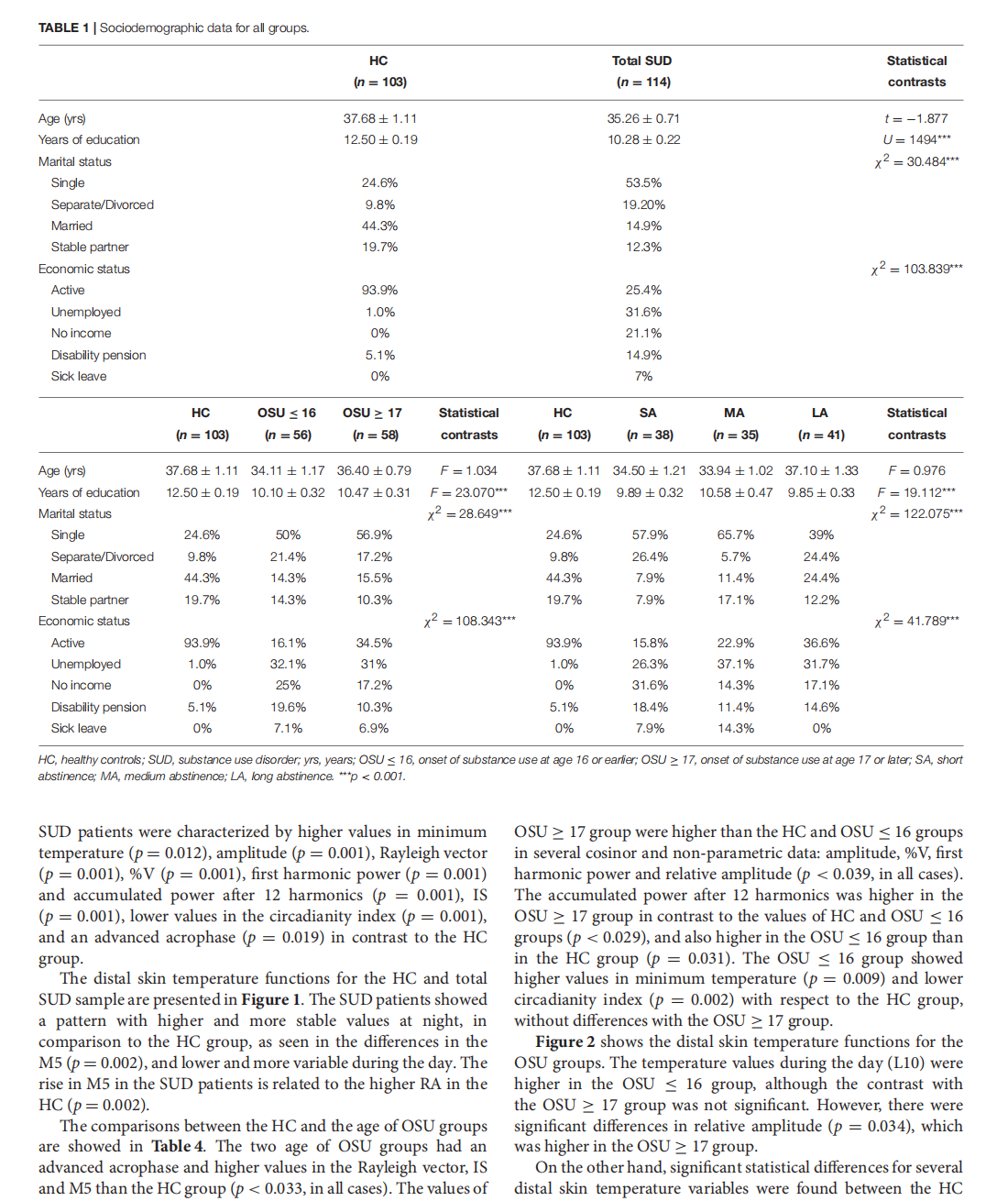
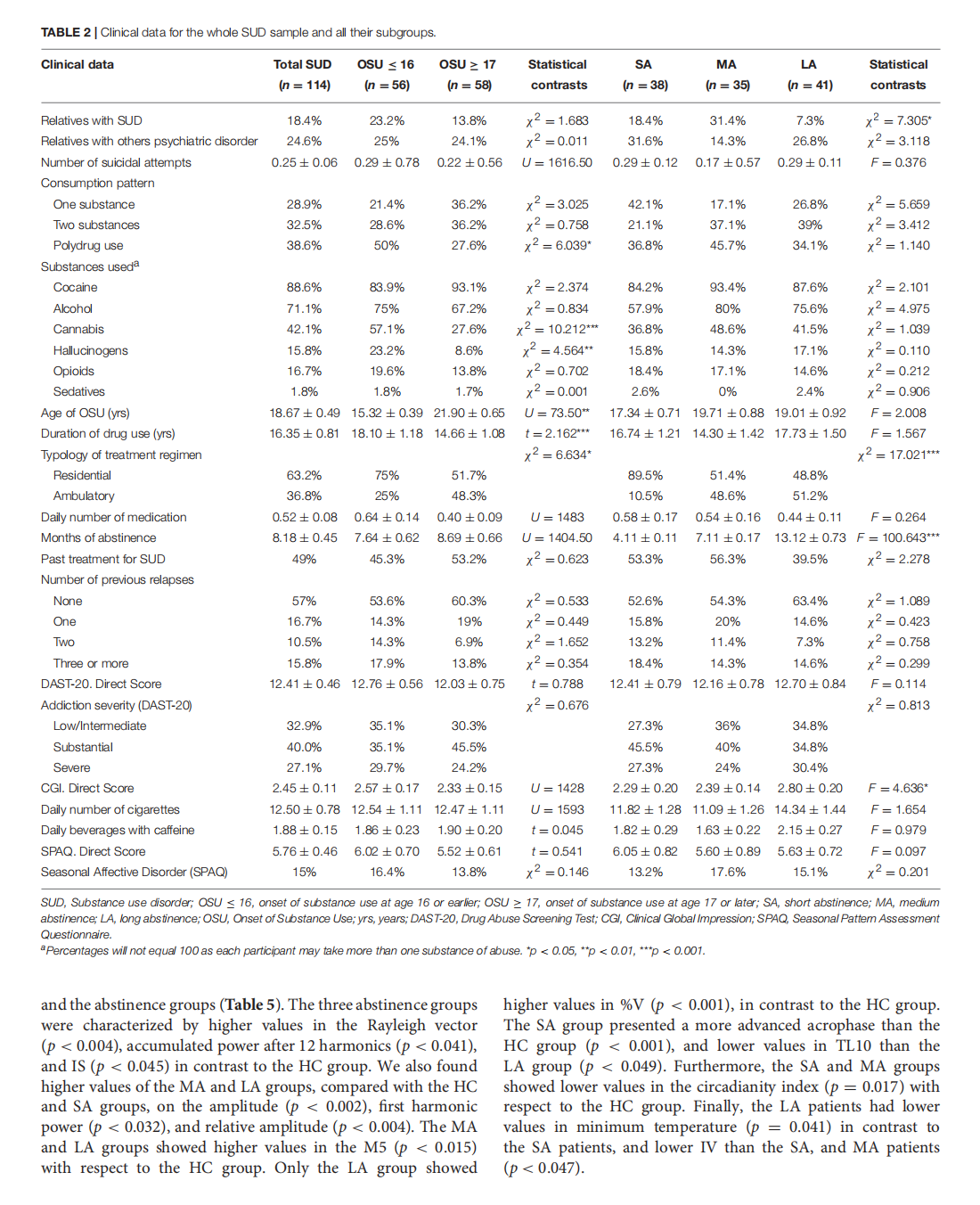
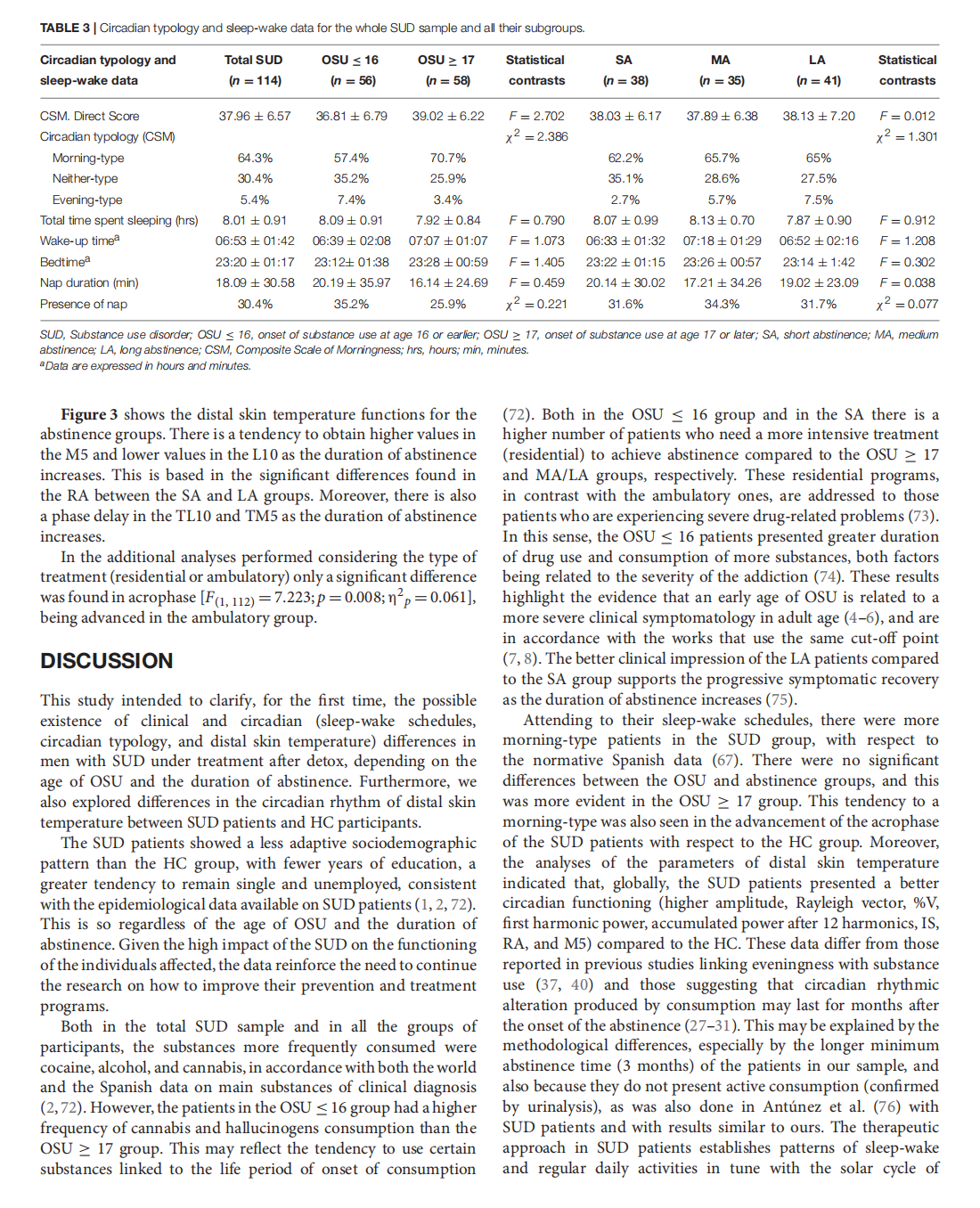
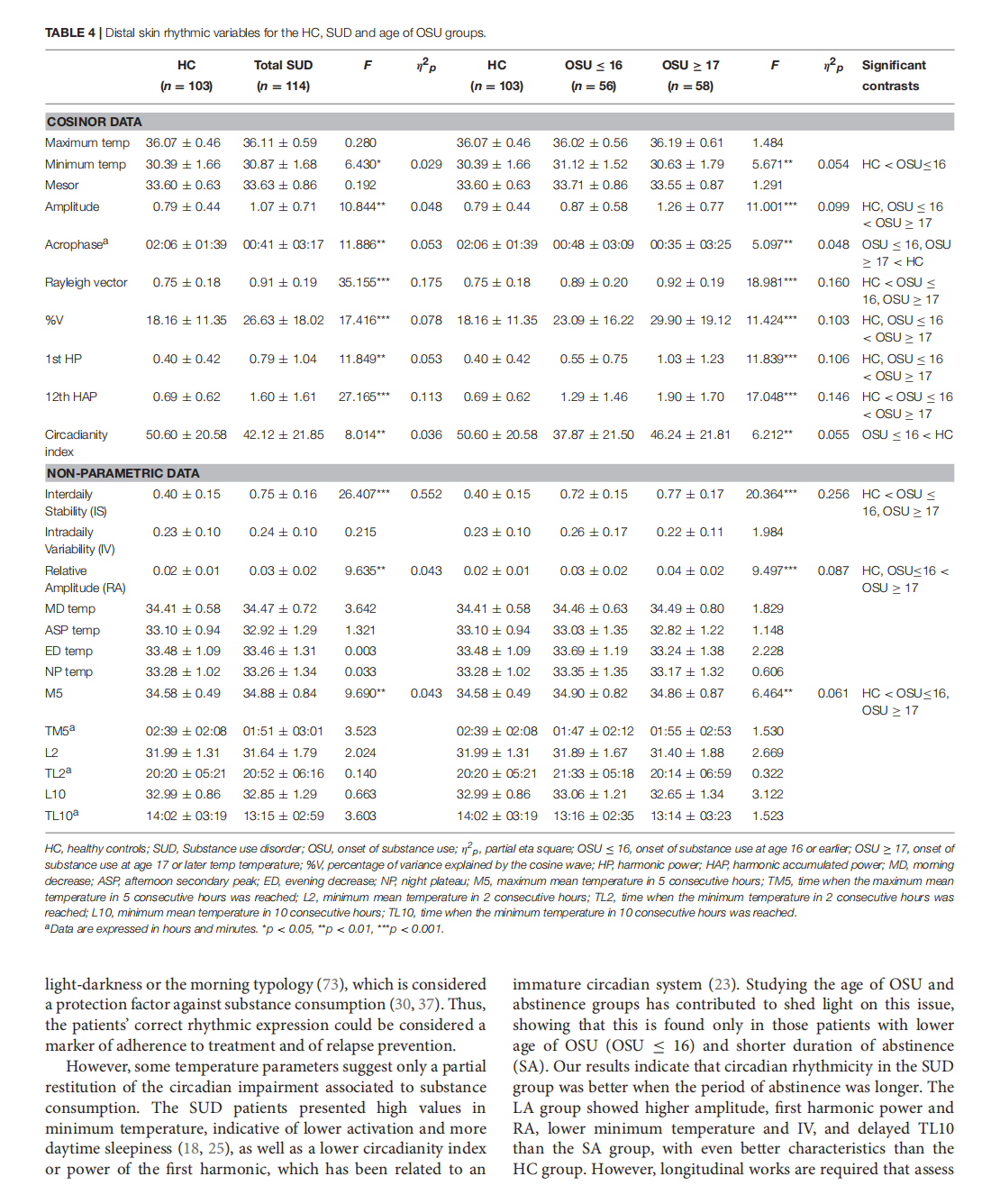
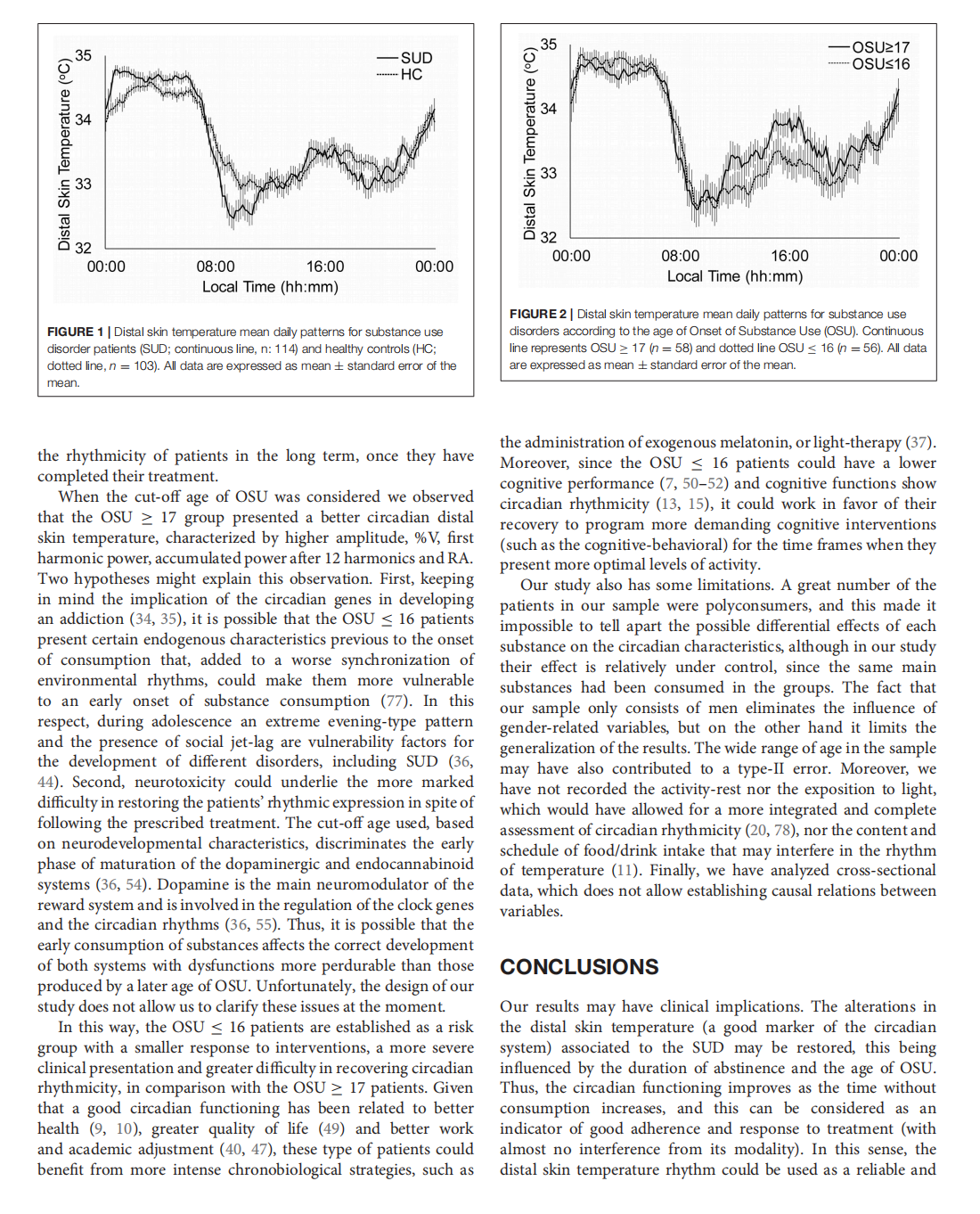
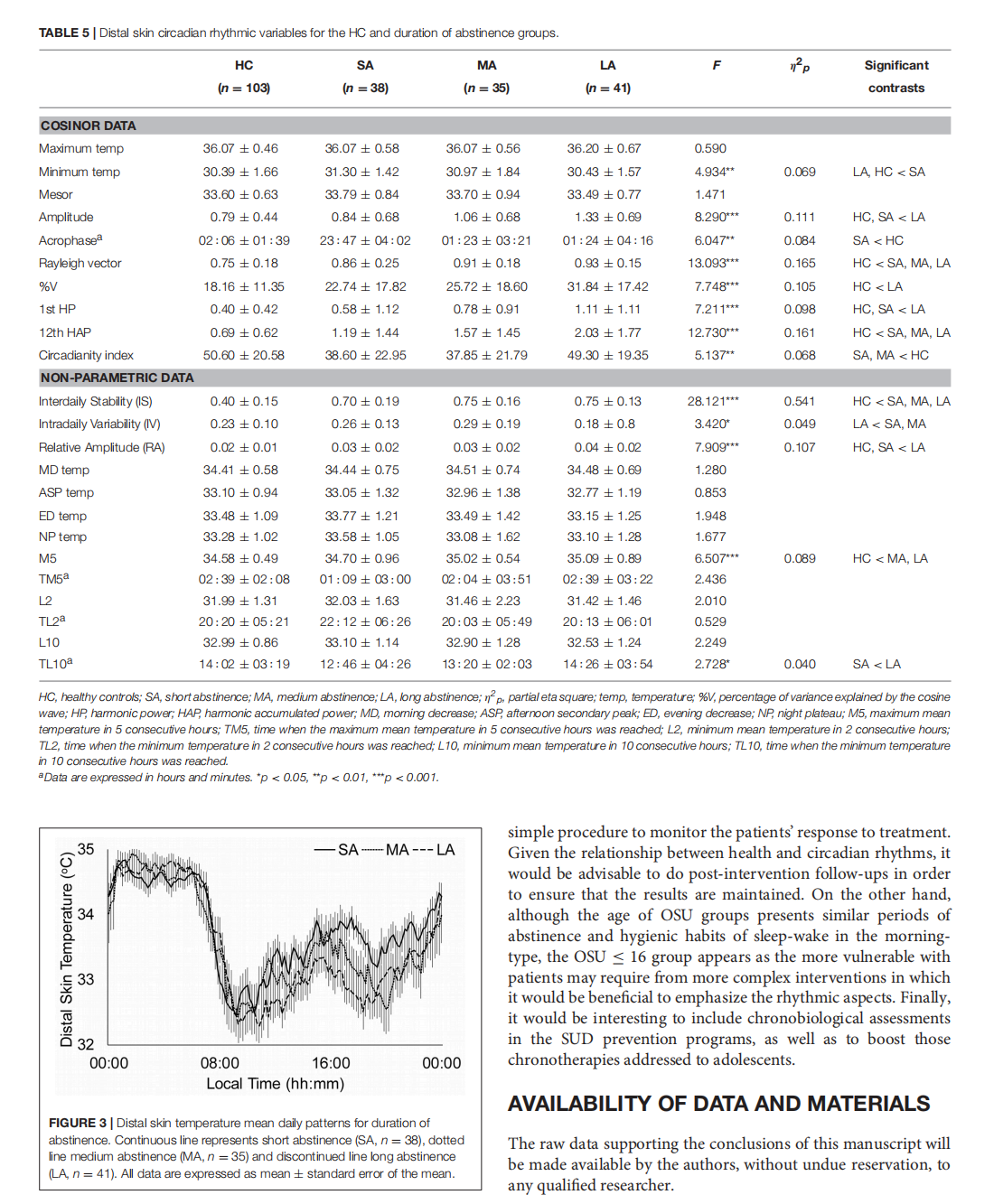



This article is excerpted from the《Frontiers in Psychiatry》 by Wound World

- 星期三, 03 12月 2025
Profiling of Petroselinum sativum (mill.) fuss phytoconstituents and assessment of their biocompatibility, antioxidant, anti-aging, wound healing, and antibacterial activities




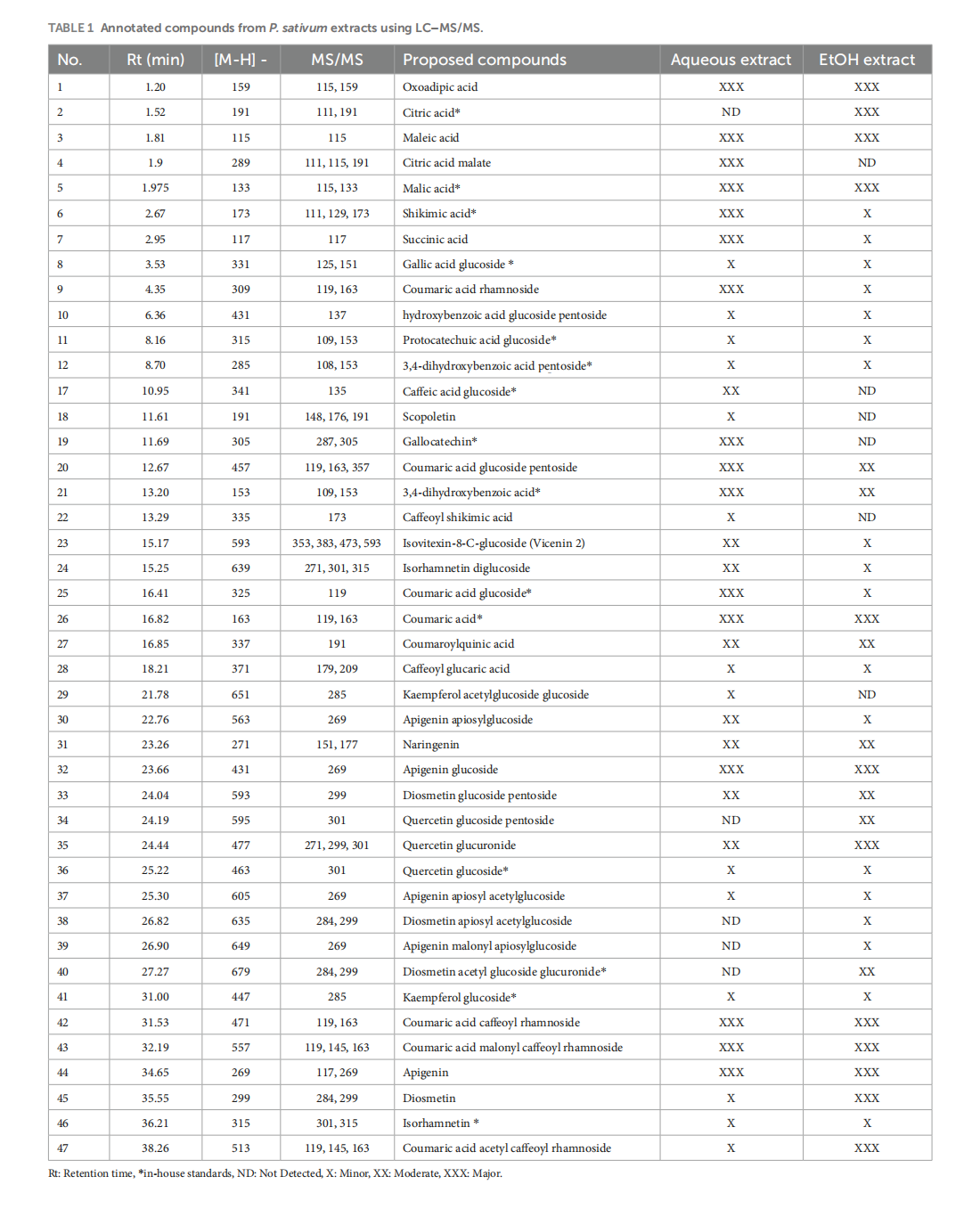
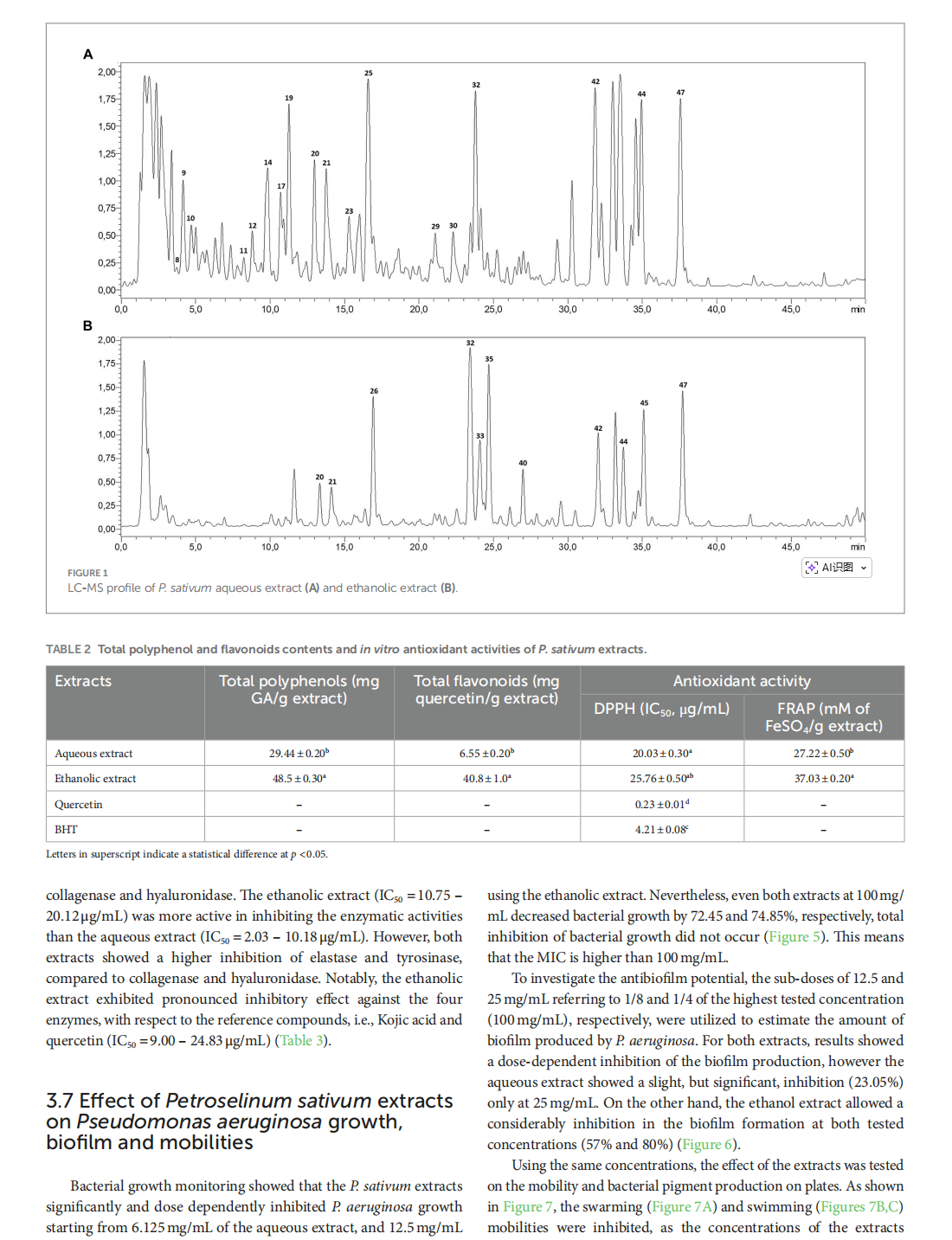

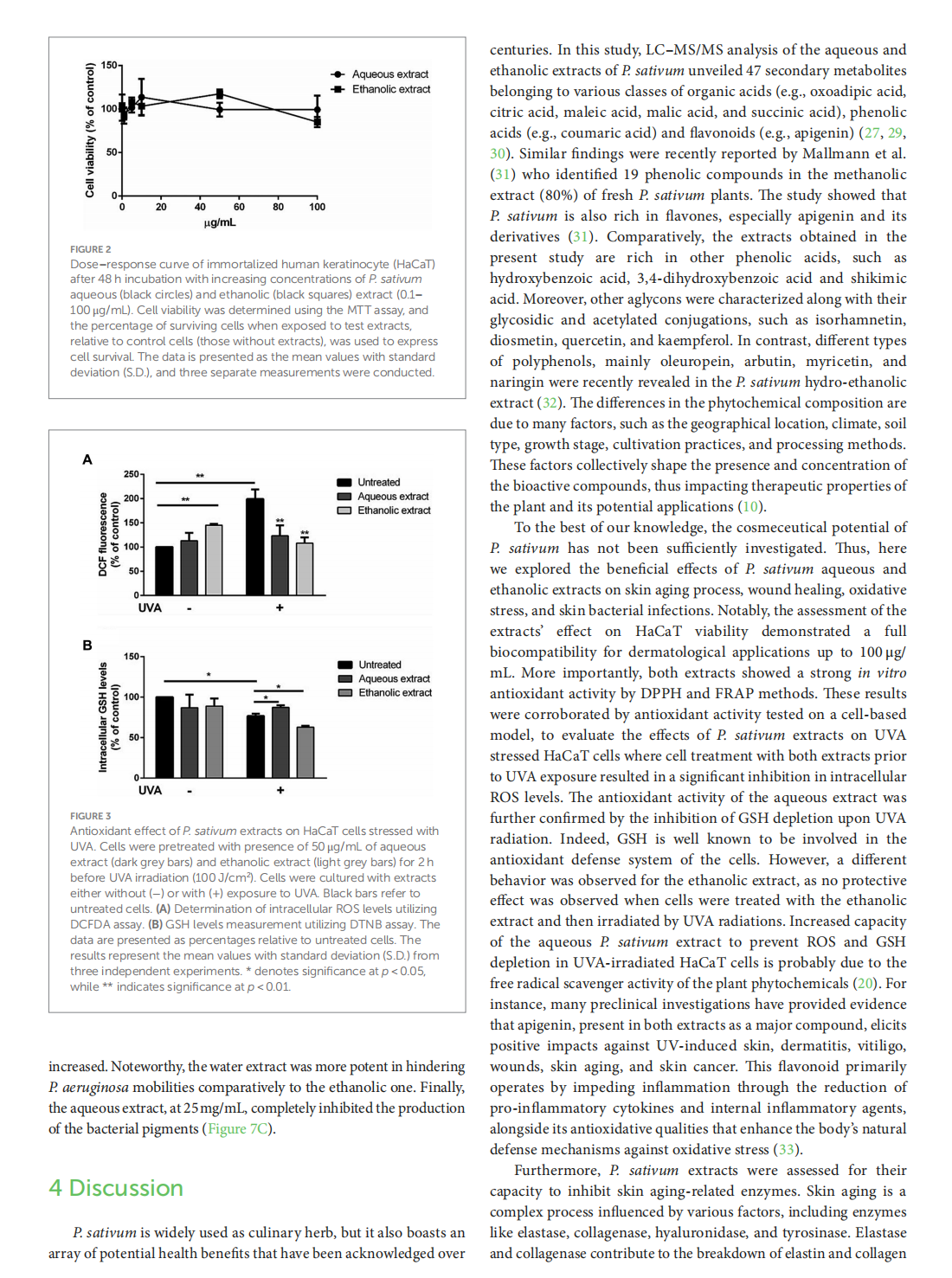
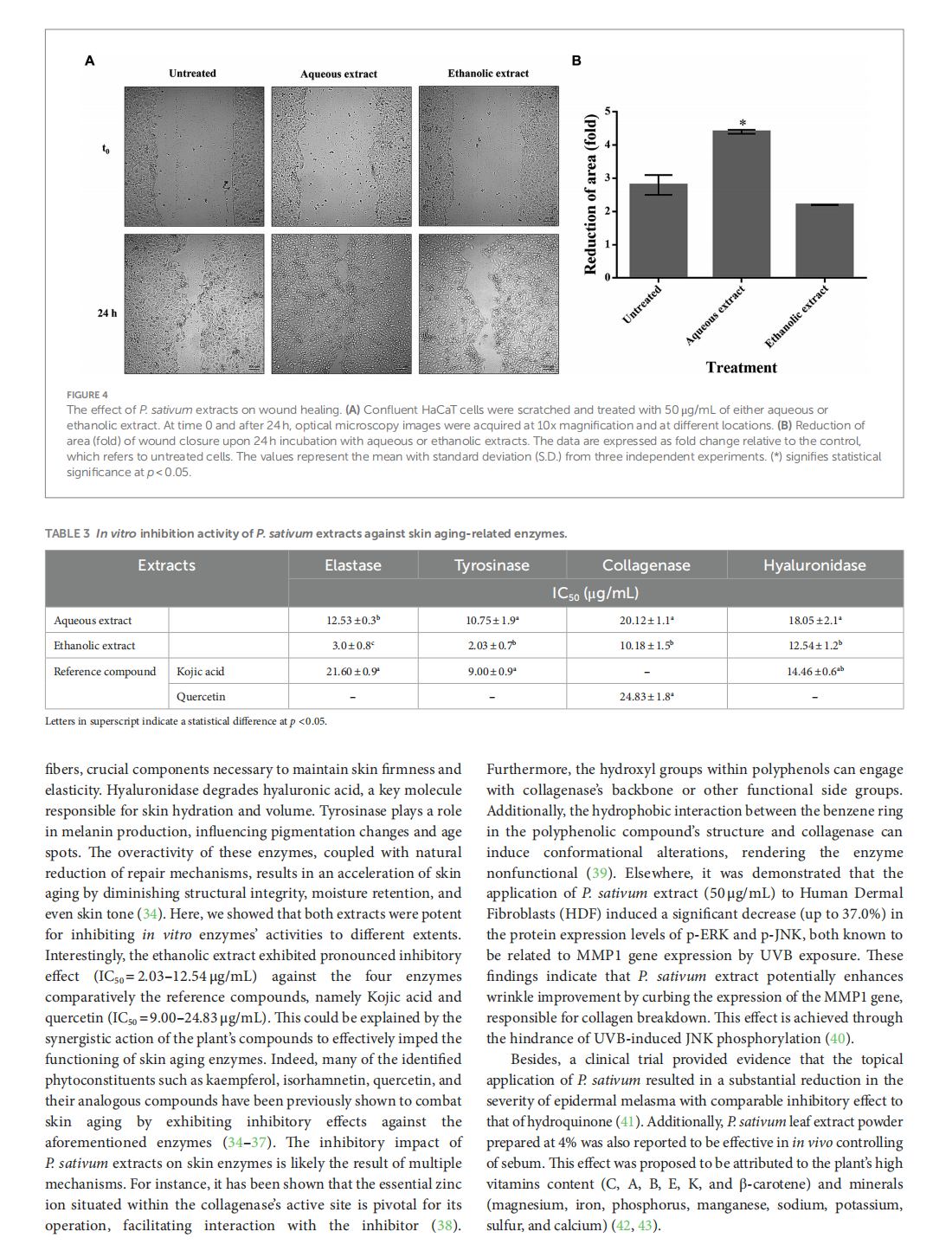
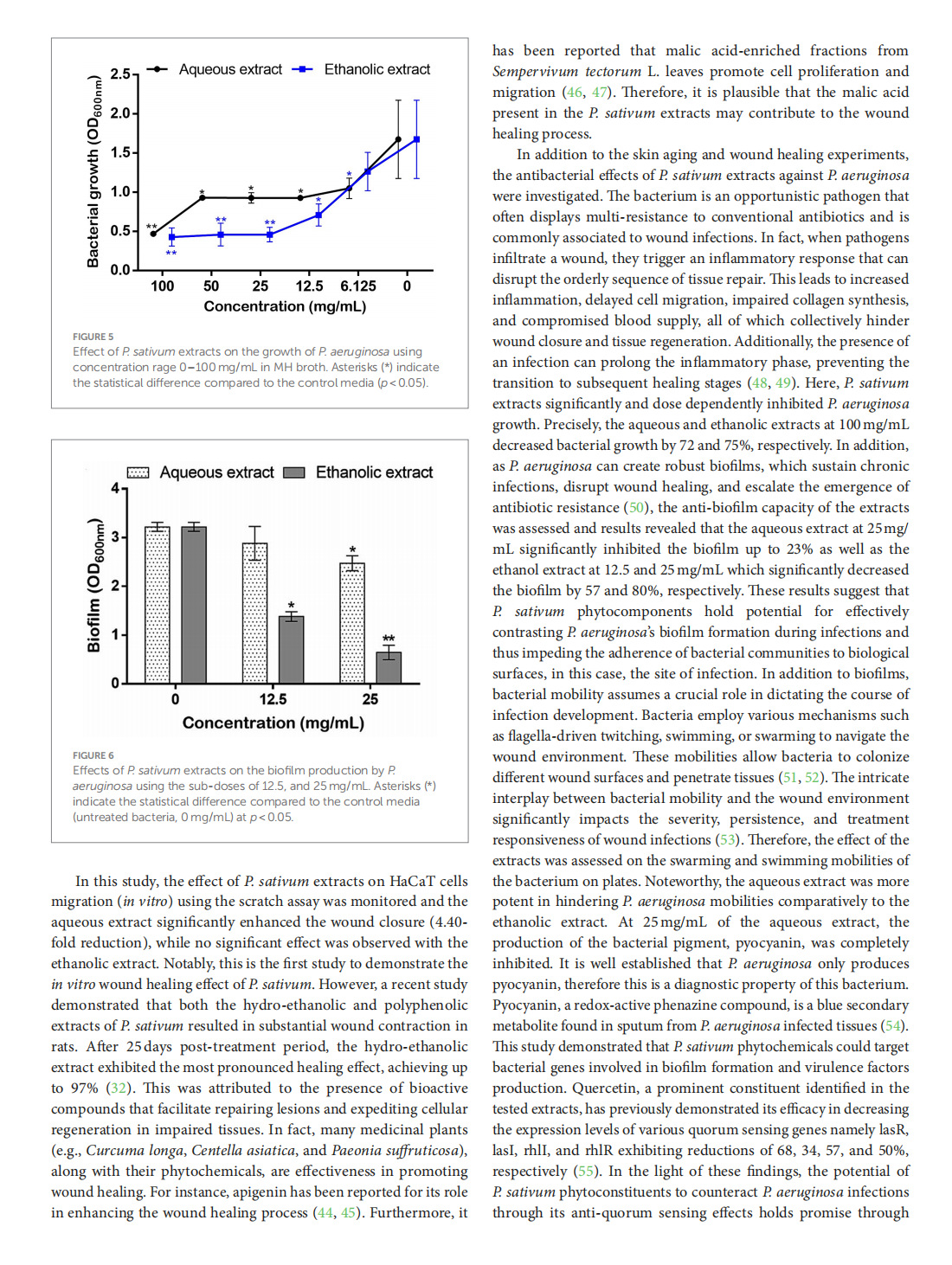
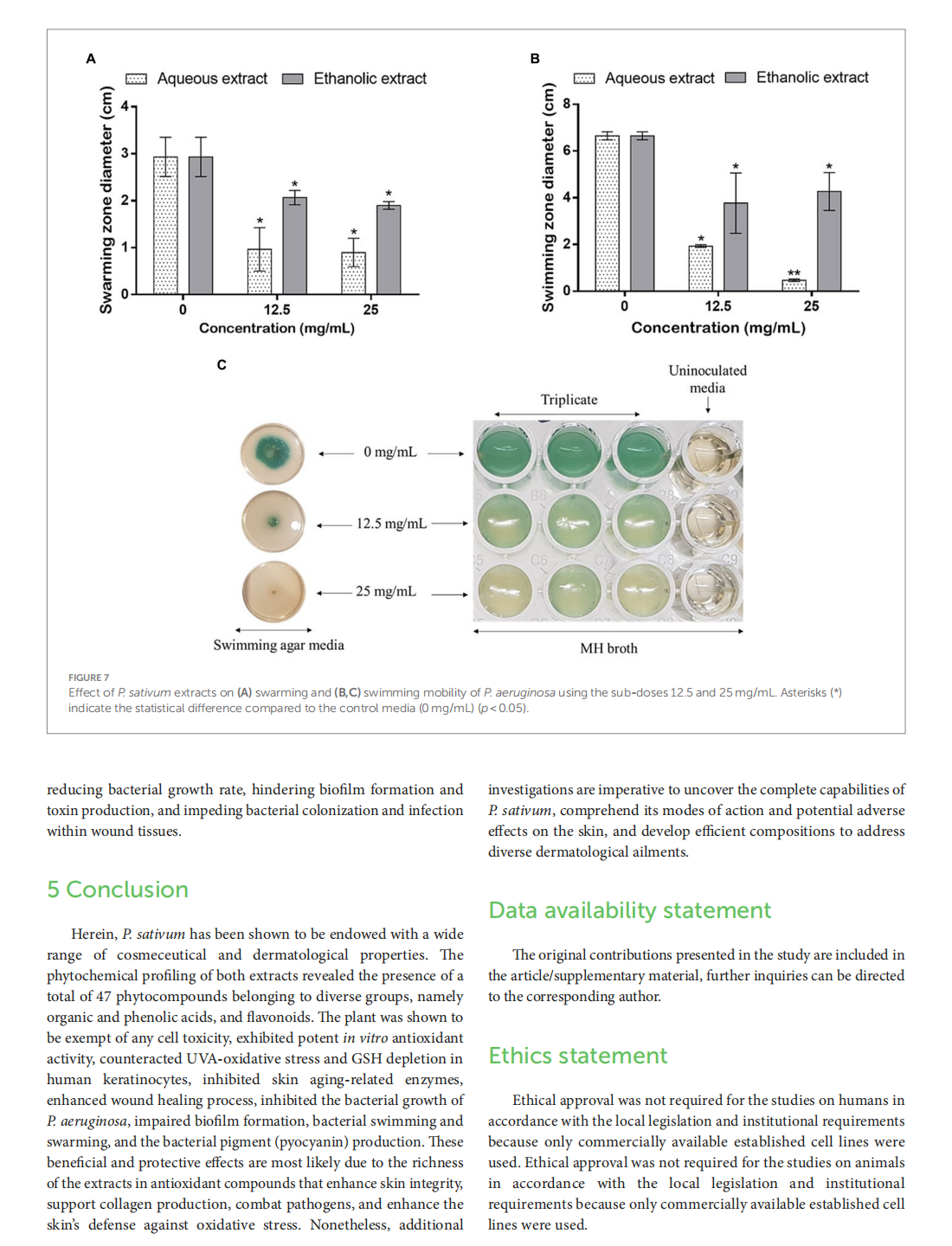


This article is excerpted from the《Frontiers in Nutrition》 by Wound World

- 星期二, 02 12月 2025
Early nociceptive evoked potentials in symptomatic and asymptomatic transthyretin mutation carriers
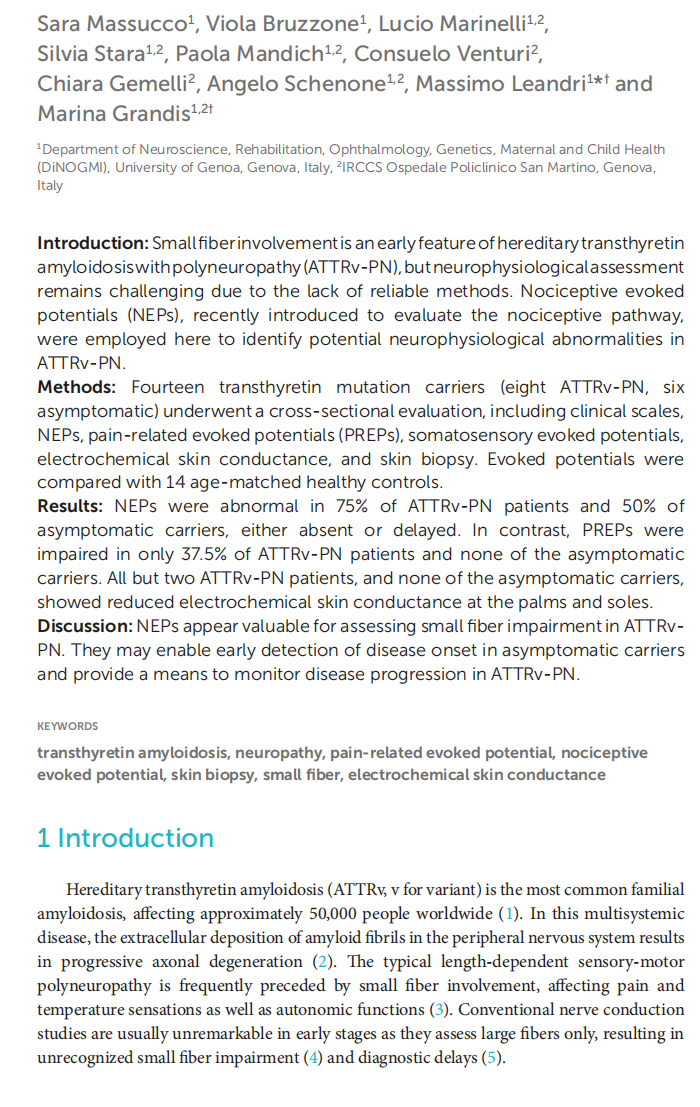



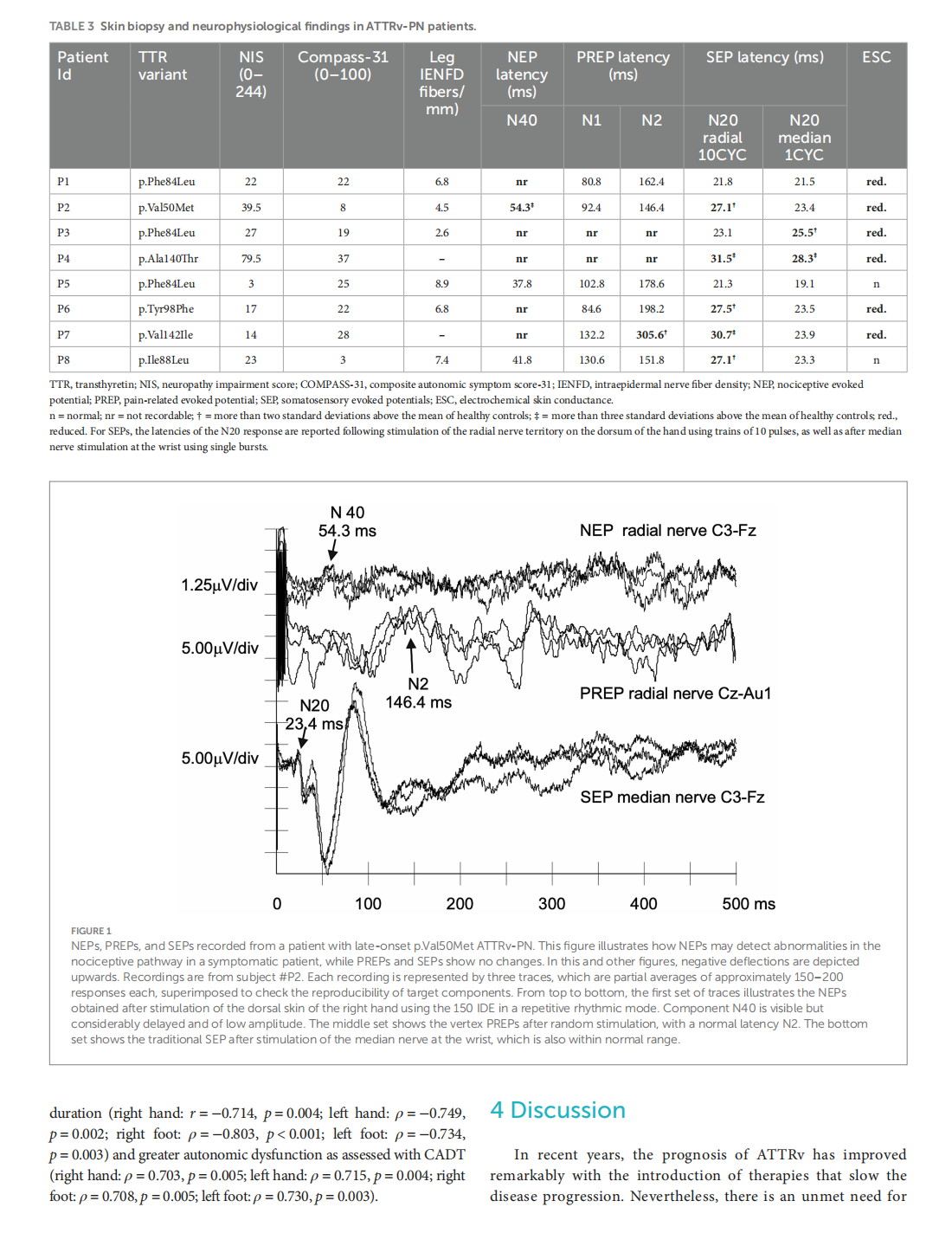
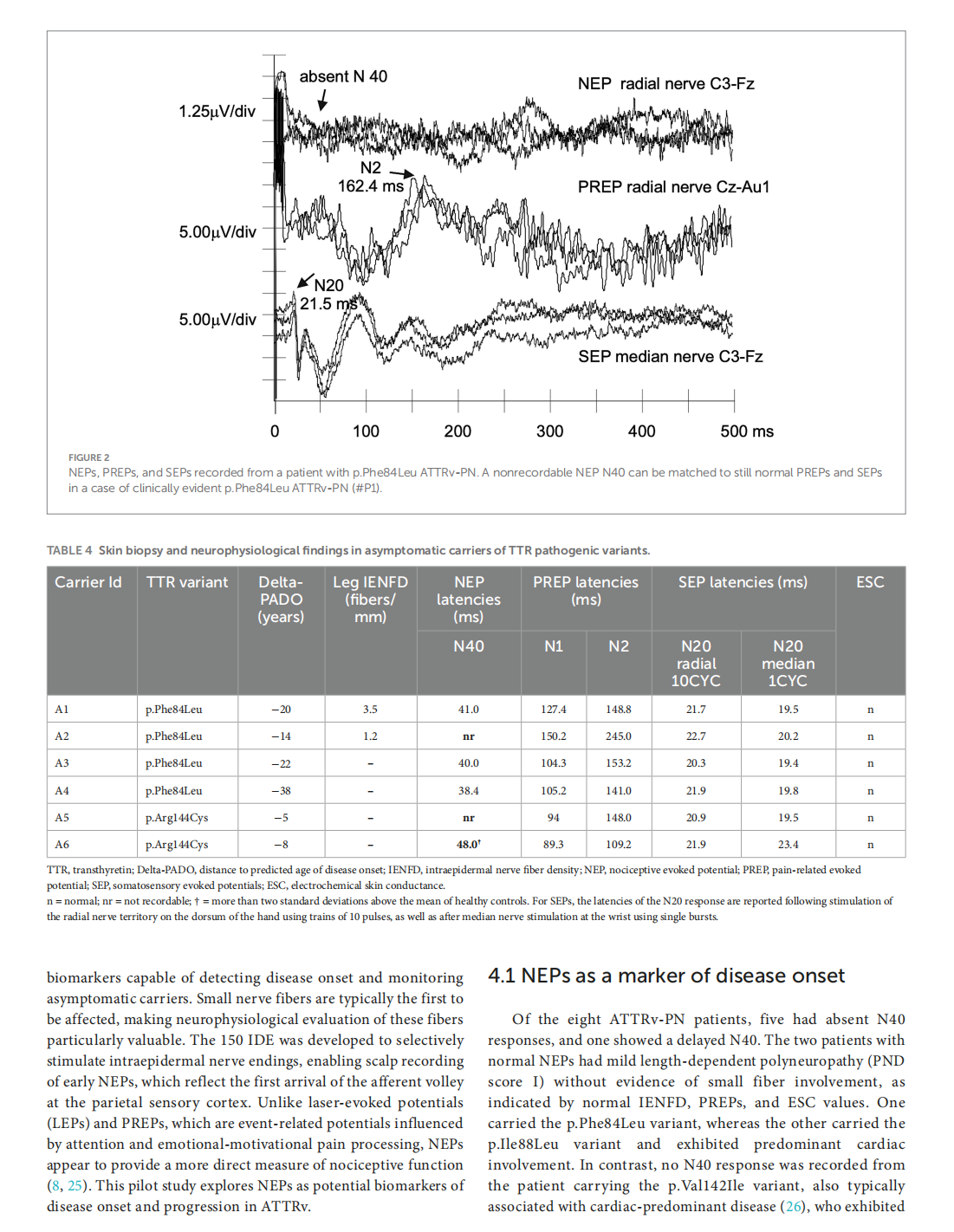



This article is excerpted from the《Frontiers in Neurology》 by Wound World

- 星期一, 01 12月 2025
Small and large fiber neuropathy in adults with myotonic dystrophy type 1

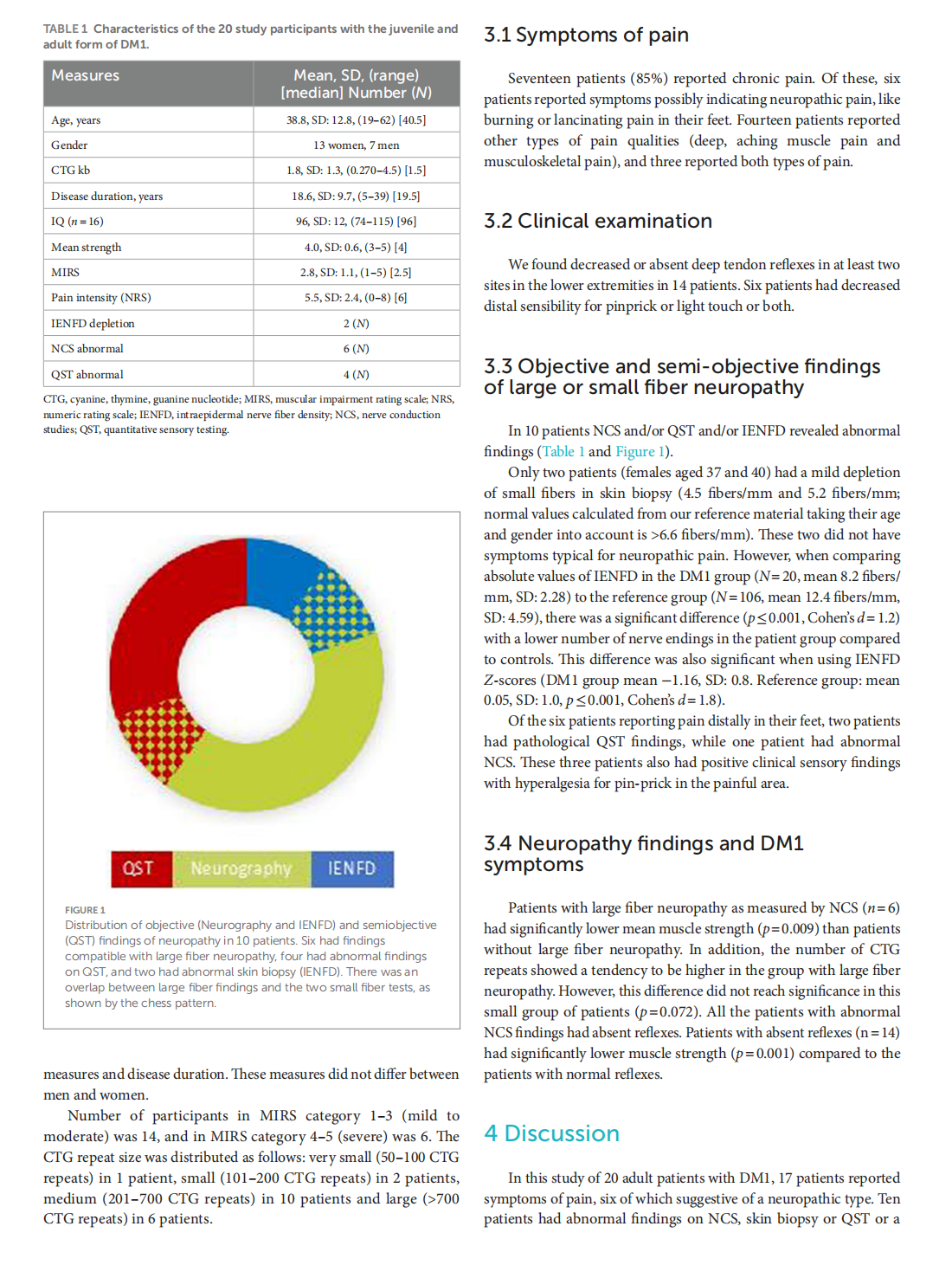


This article is excerpted from the《Frontiers in Neurology》 by Wound World

- 星期五, 28 11月 2025
Which Method for Diagnosing Small Fiber Neuropathy?



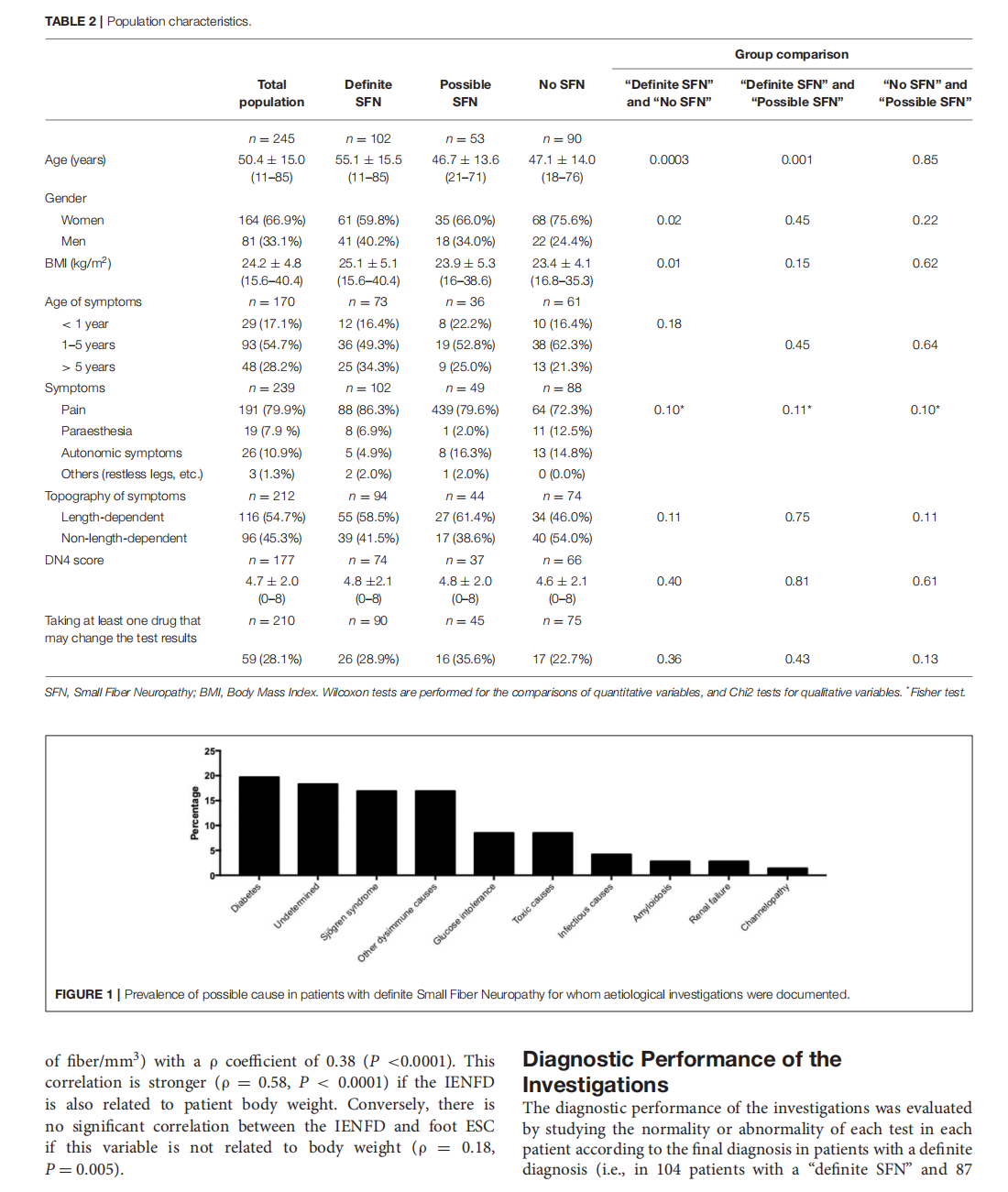
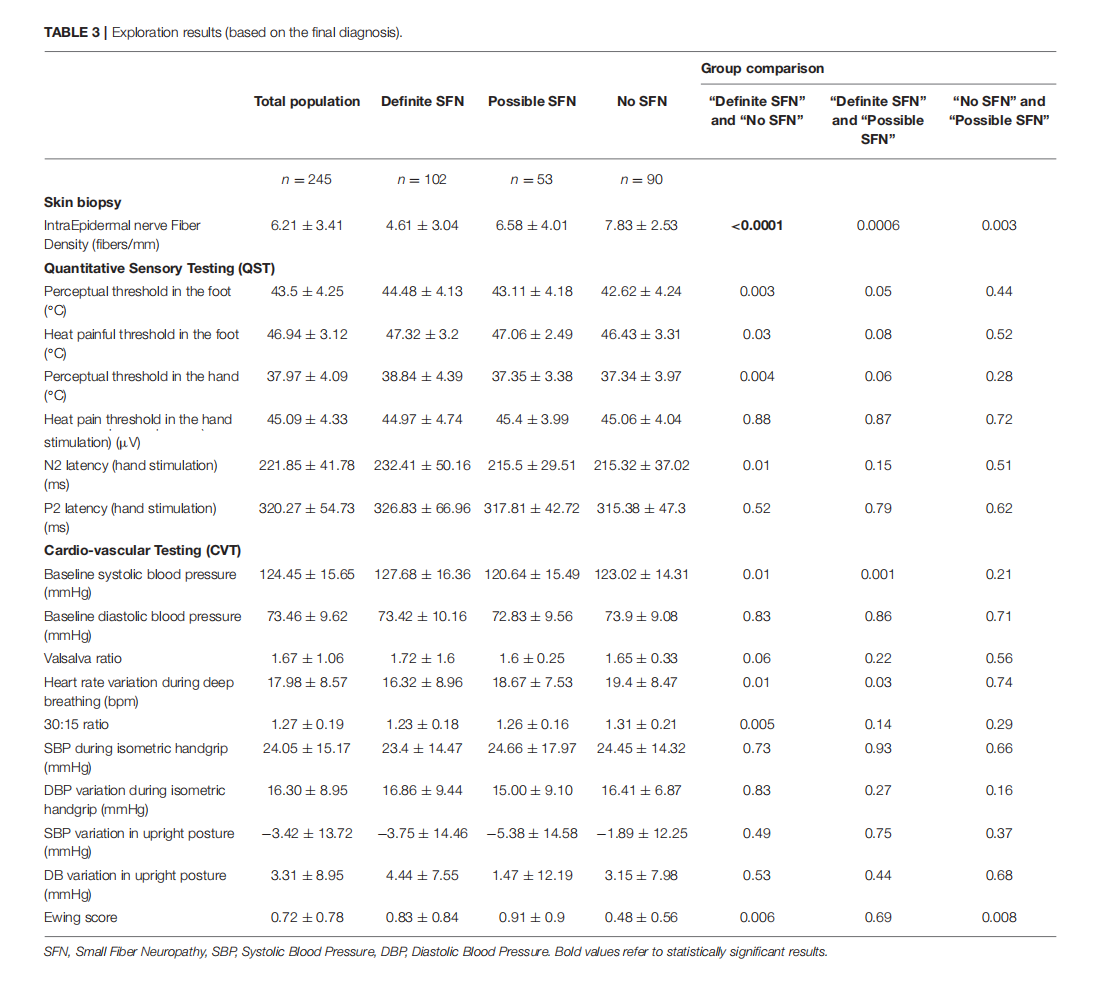
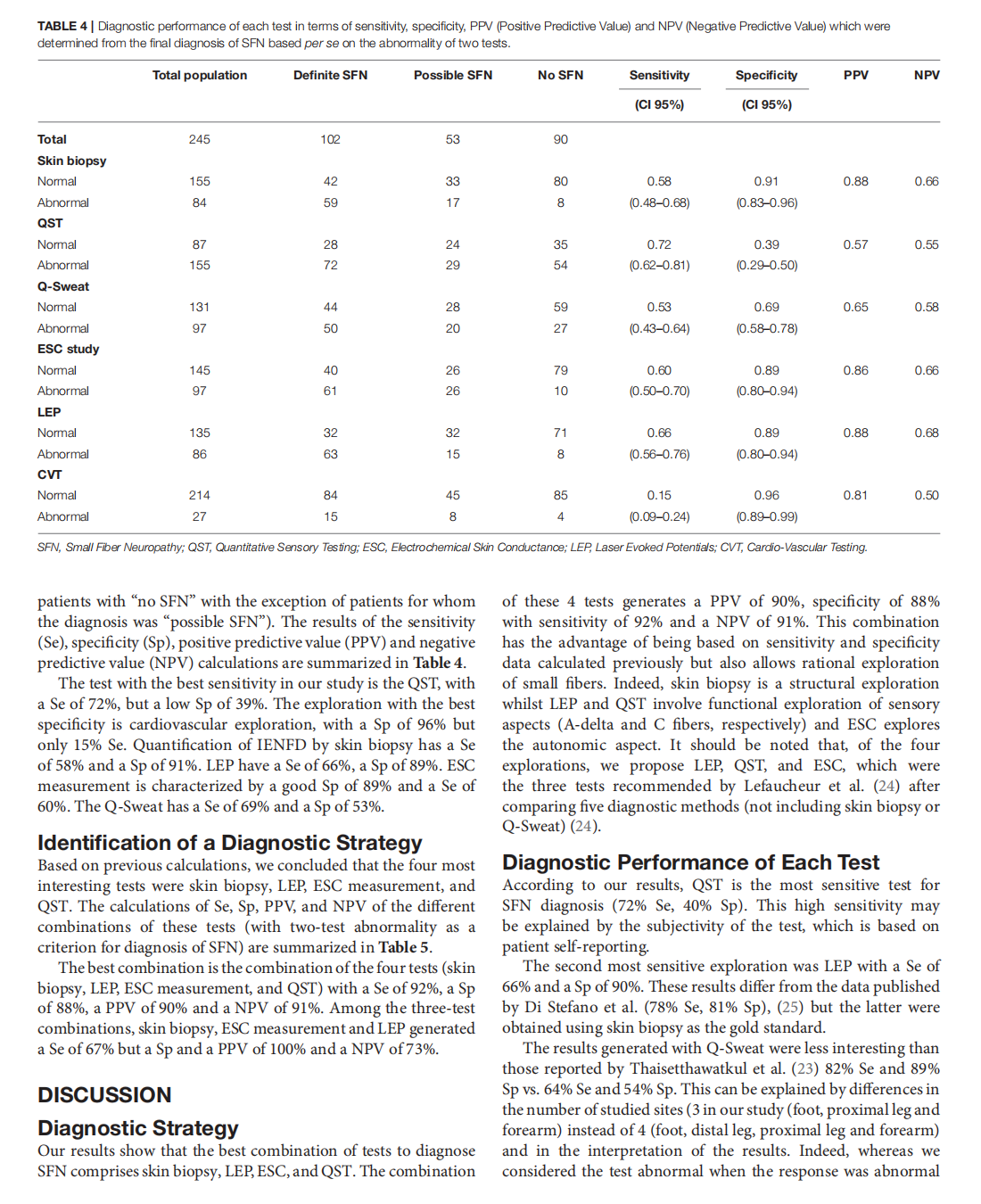



This article is excerpted from the《Frontiers in Neurology》 by Wound World

- 星期四, 27 11月 2025
Getting under the skin of the menopausal hot flush: a protocol to examine skin function and structure in symptomatic postmenopausal women


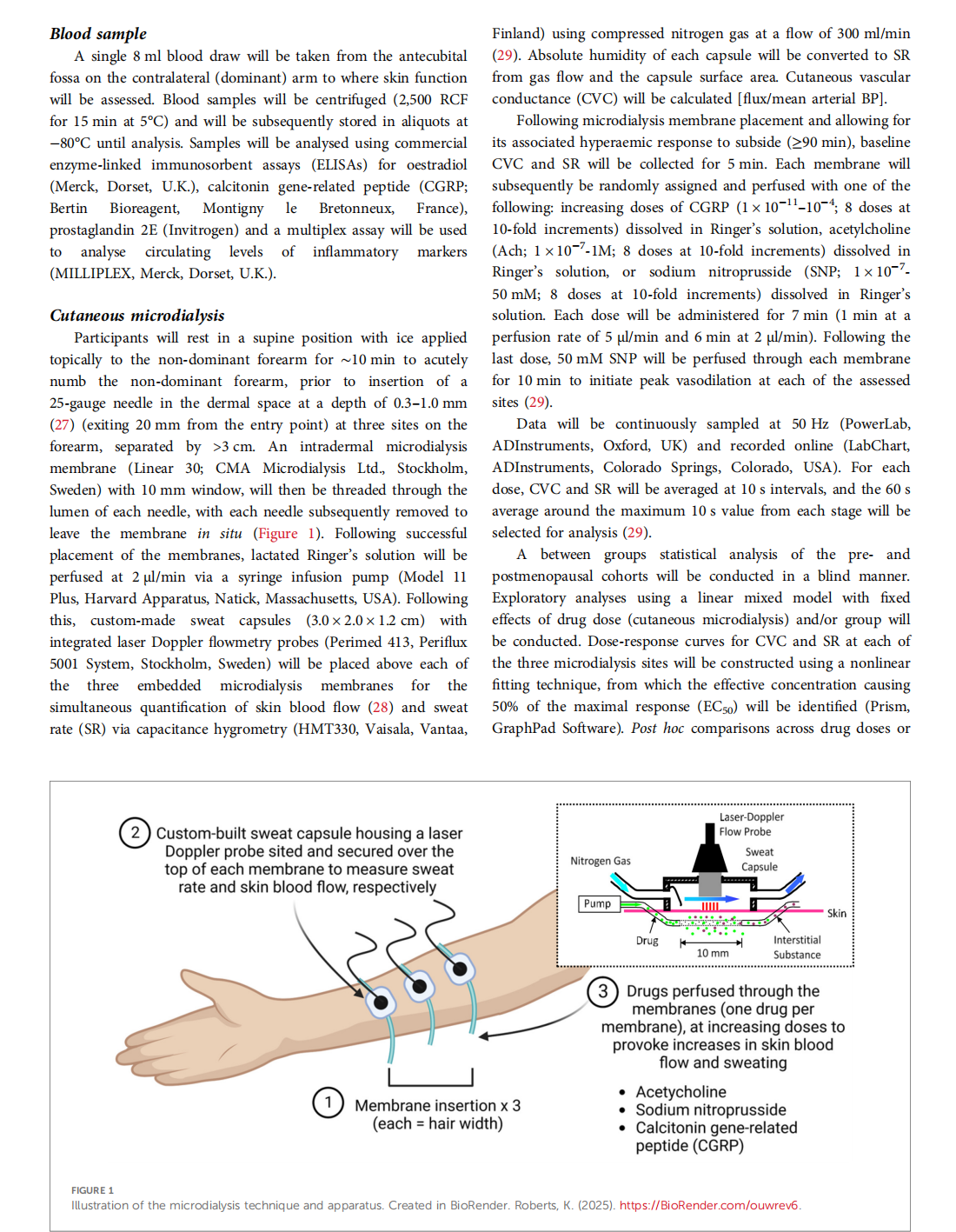



This article is excerpted from the 《Frontiers in Global Women's Health》 by Wound World
