
















This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Javier Ganz,1,2,3,8,9 Lovelace J. Luquette,4,8 Sara Bizzotto,1,2,3,5,8 Michael B. Miller,1,3,6 Zinan Zhou,1,2,3 Craig L. Bohrson,4
Hu Jin,4 Antuan V. Tran,4 Vinayak V. Viswanadham,4 Gannon McDonough,6 Katherine Brown,6 Yasmine Chahine,1
Brian Chhouk,1 Alon Galor,4 Peter J. Park,4,7,* and Christopher A. Walsh1,2,3,10,*
1 Division of Genetics and Genomics, Manton Center for Orphan Disease Research, Department of Pediatrics, and Howard Hughes Medical Institute, Boston Childrens Hospital, Boston, MA 02115, USA
2 Departments of Pediatrics and Neurology, Harvard Medical School, Boston, MA 02115, USA
3 Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA
4 Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA
5 Sorbonne Universite´ , Institut du Cerveau (Paris Brain Institute) ICM, Inserm, CNRS, Hoˆ pital de la Pitie´ Salpeˆ trie`re, 75013 Paris, France
6 Department of Pathology, Brigham and Womens Hospital, Harvard Medical School, Boston, MA 02115, USA
7 Division of Genetics, Brigham and Womens Hospital, Boston, MA 02115, USA
8 These authors contributed equally
9 Present address: Merck Research Laboratories, Cambridge, MA 02142, USA
10 Lead contact
*Correspondence: 该Email地址已收到反垃圾邮件插件保护。要显示它您需要在浏览器中启用JavaScript。 (P.J.P.), 该Email地址已收到反垃圾邮件插件保护。要显示它您需要在浏览器中启用JavaScript。 (C.A.W.)
https://doi.org/10.1016/j.cell.2024.02.025
SUMMARY
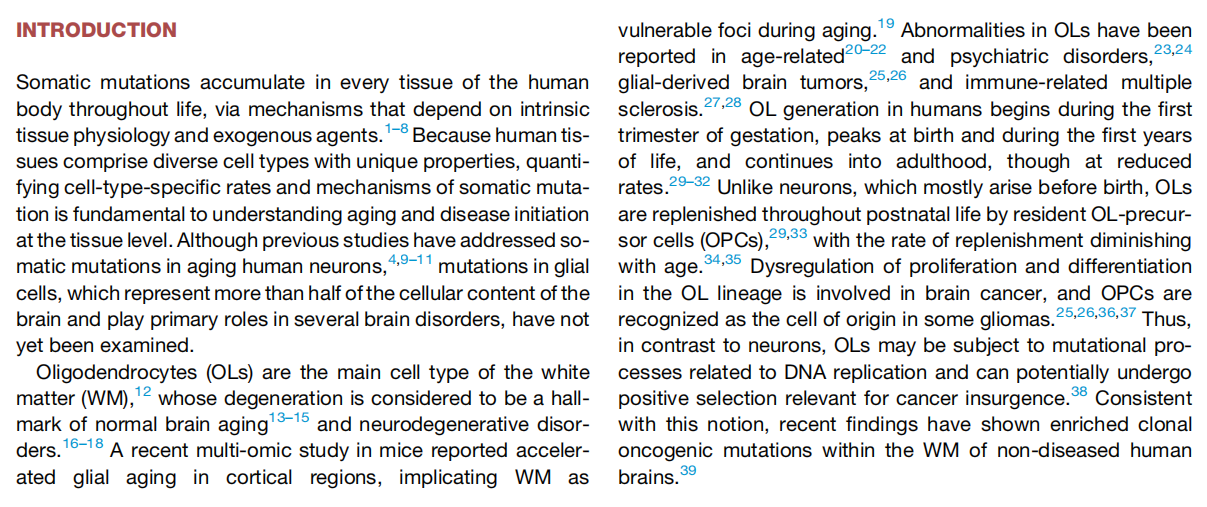
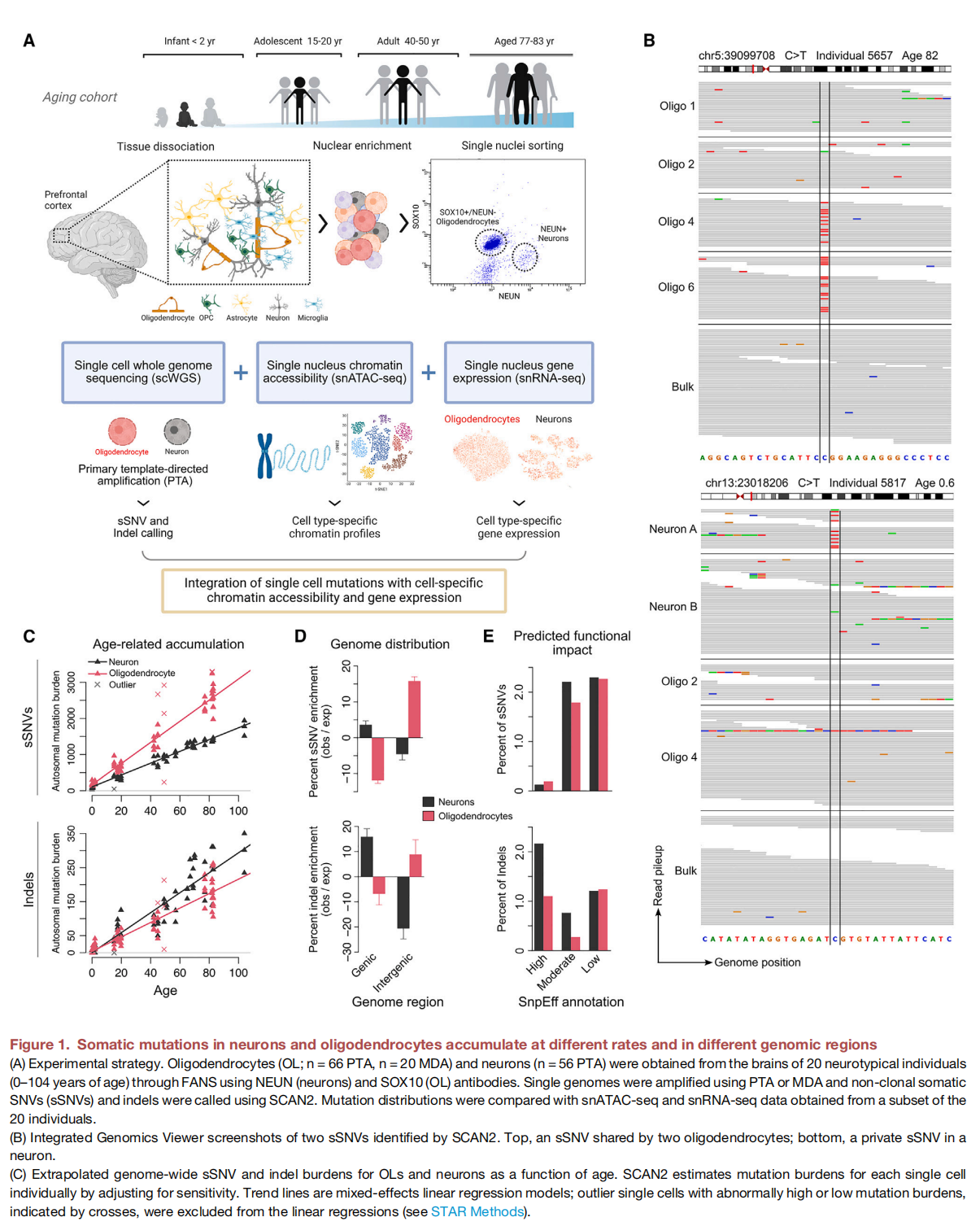
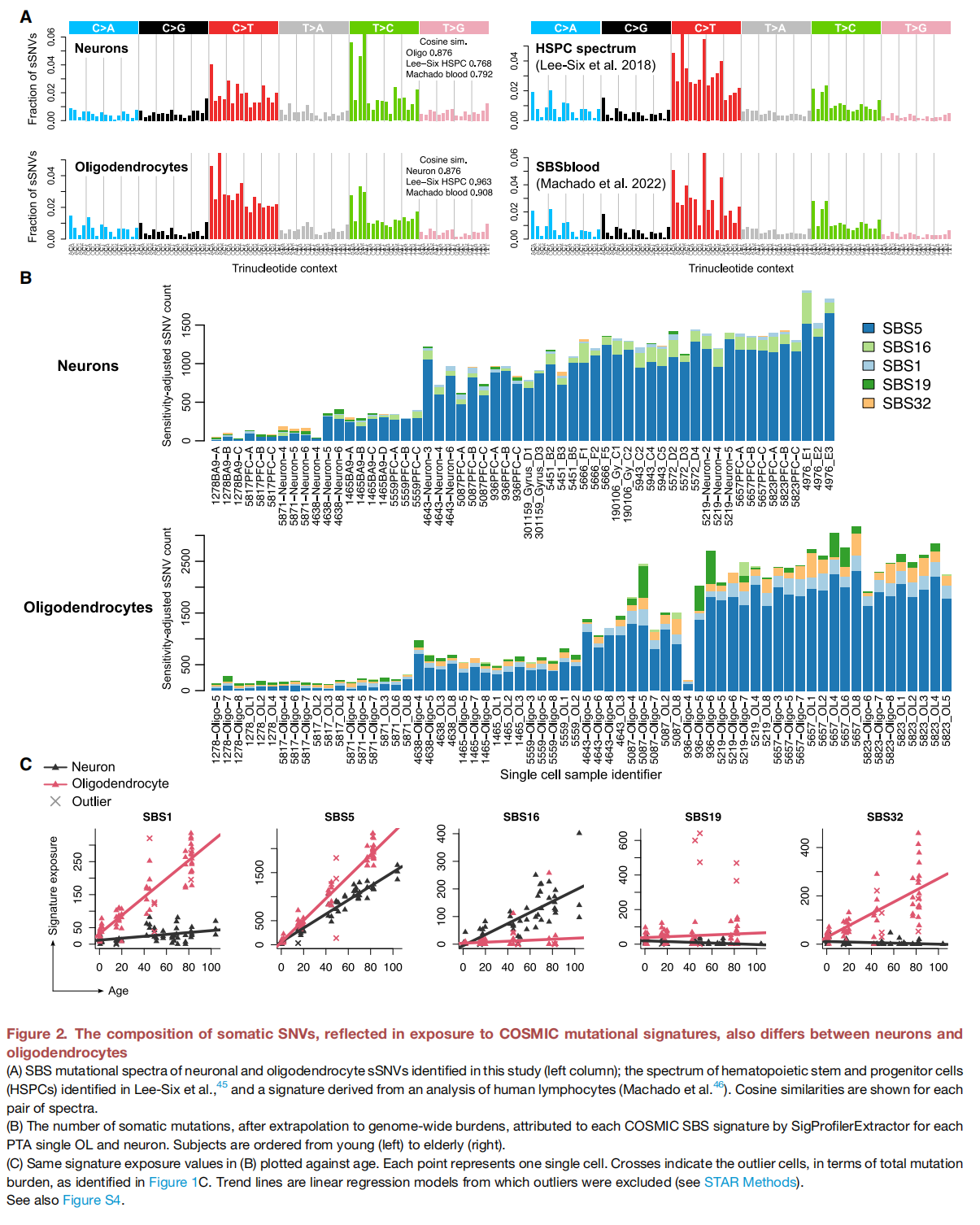
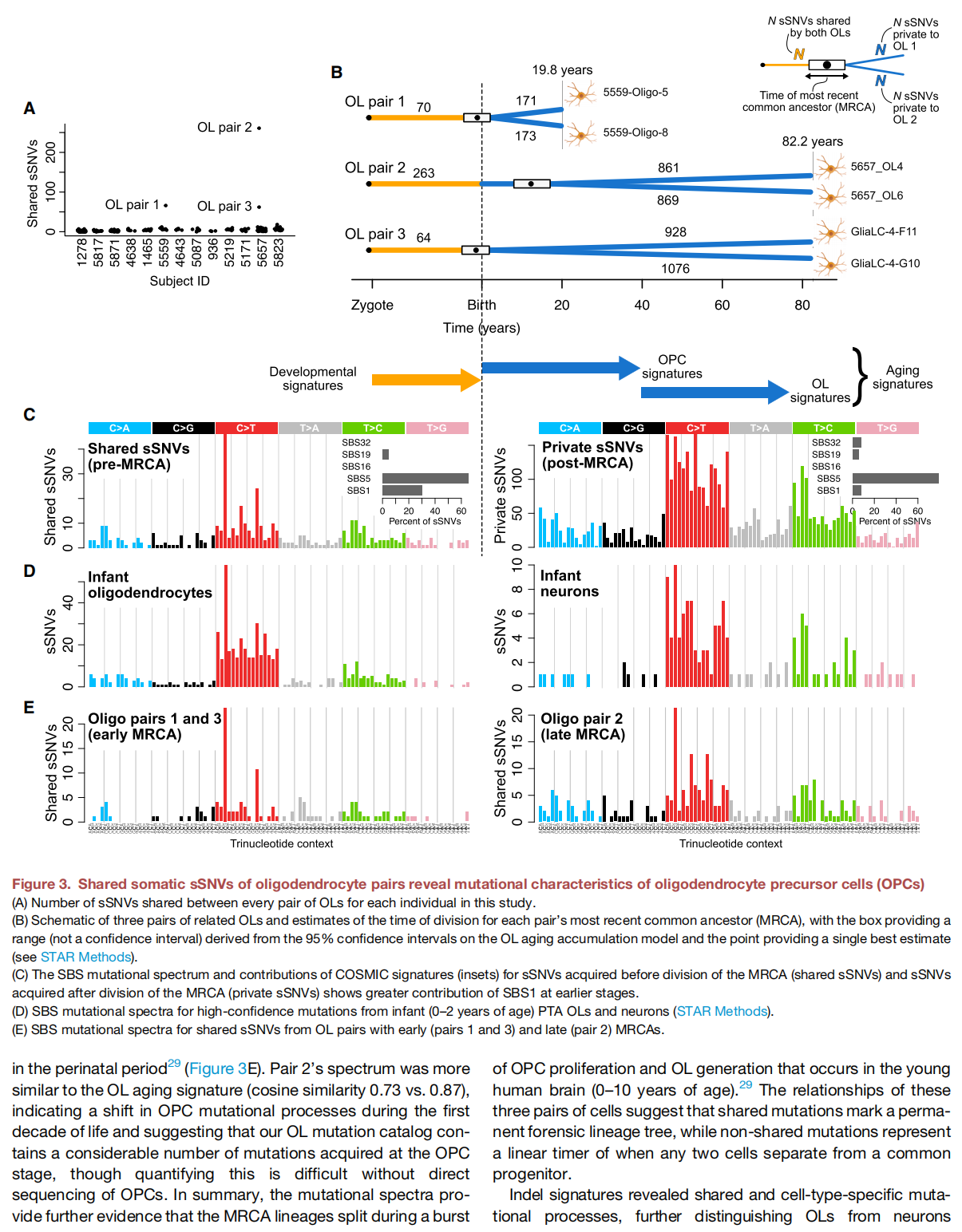
Characterizing somatic mutations in the brain is important for disentangling the complex mechanisms of aging, yet little is known about mutational patterns in different brain cell types. Here, we performed wholegenome sequencing (WGS) of 86 single oligodendrocytes, 20 mixed glia, and 56 single neurons from neurotypical individuals spanning 0.4–104 years of age and identified >92,000 somatic single-nucleotide variants (sSNVs) and small insertions/deletions (indels). Although both cell types accumulate somatic mutations linearly with age, oligodendrocytes accumulated sSNVs 81% faster than neurons and indels 28% slower than neurons. Correlation of mutations with single-nucleus RNA profiles and chromatin accessibility from the same brains revealed that oligodendrocyte mutations are enriched in inactive genomic regions and are distributed across the genome similarly to mutations in brain cancers. In contrast, neuronal mutations are enriched in open, transcriptionally active chromatin. These stark differences suggest an assortment of active mutagenic processes in oligodendrocytes and neurons.







































This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
